|
|
|
[
]
2006 [
] [
] [
] [
] [
]

|
2006 ANNUAL RESEARCH REVIEW Poster Session (Oct, 2006)
Stereo Vision-aided Navigation for Robotic Boats,
Arvind Menezes Pereira, Gabriel Sibley, Gaurav Sukhatme, Amit Dhariwal, Bin Zhang, Carl Oberg, Beth Stauffer, Stefanie Moorthi and David Caron.
|
|

|
International Conference on Distributed Computing in Sensor Systems(DCOSS) (Jun 2006)
The First ACM International Workshop on UnderWater Networks (WUWNet 06)(Sep 2006)
2006 ANNUAL RESEARCH REVIEW Poster Session (Oct, 2006)
NAMOS: Networked Aquatic Microbial Observing System,
Amit Dhariwal, Bin Zhang, Arvind Pereira, Carl Oberg, Beth Stauffer, Stefanie Moorthi, David Caron and Gaurav Sukhatme
As part of our research, we have designed and developed sensor-actuated network for marine monitoring. The network consists of ten static buoy nodes and one mobile
robotic boat for real-time in-situ measurements and analysis of chemical, physical and biologically pertinent phenomena governing the abundance of micro-organisms at
relevant spatio-temporal scales. The goal of the network is to obtain high-resolution information on the spatial and temporal distribution of plankton assemblages in
aquatic environments using the in situ presence afforded by the network, and to make possible network-enabled robotic sampling of hydrographic features of interest.
|
|

|
American Society of Limnology and Oceanography (ASLO) (Jun, 2006)
Observation of Phytoplankton Bloom Dynamics in a Small Lake Using a Wireless Sensor Network,
Beth A. Stauffer, David A. Caron, Gaurav S. Sukhatme, Bin Zhang, Amit Dhariwal, Carl Oberg, Stefanie D. Moorthi, Arvind Menezes-Pereira, William Kaiser, Mark Hansen, Maxim Batalin
Effective monitoring and detection of harmful blooms of algae and cyanobacteria require the collection, analysis, and modeling of large sets of data collected from susceptible environments.
The use of embedded sensor networks to define important environmental features or events and direct sample collection to areas of greatest concern or interest can significantly increase the
efficiency of sensing and sampling strategies. To this end, the Networked Aquatic Microbial Observing System (NAMOS) has been developed, consisting of mobile and static sensing platforms,
which cooperatively direct sampling and higher resolution sensing to areas of biological interest. A complementary robotic platform, the Networked Infomechanical System (NIMS), has also
been employed in the study of plankton dynamics in aquatic ecosystems.
|
|

|
2006 NSF SITE VISIT Poster Session (Jun, 2006)
Multi-scale Actuation and Sensing: An Overview,
Bill Kaiser, Mark Hansen, Gaurav Sukhatme and the MAS Team
|
|

|
2006 NSF SITE VISIT Poster Session (Jun, 2006)
ENS Approaches for Observing and Sensing of Microbial Communities in Aquatic Ecosystems An Overview,
Gaurav Sukhatme, David Caron, Carl Oberg, Bin Zhang, Amit Dhariwal, Beth Stauffer, Stefanie Moorthi, and Arvind Pereira
|
|

|
2006 NSF SITE VISIT Poster Session (Jun, 2006)
Characterization of the Phytoplankton Community in Lake Fulmor, CA, Using Embedded Sensor Networks ,
Beth Stauffer, Stefanie Moorthi, Bin Zhang, Amit Dhariwal, Carl Oberg, Arvind Pereira, David Caron, and Gaurav Sukhatme
The Networked Aquatic Microbial Observing System (NAMOS) is an array of embedded networked sensors that has been developed to study plankton populations in marine and
aquatic environments. The application of NAMOS in Lake Fulmor, adjacent to the James Mountain Reserve, California, has begun to yield valuable insights into plankton
community structure and dynamics, including phytoplankton spatial and temporal distributions throughout the lake. Data collected during deployments of the system in
May, July, and October, 2005 revealed decreases in water column temperature stratification, accompanied by overall increases in chlorophyll in the lake throughout the
three deployments. Chlorophyll concentrations (indicative of phytoplankton biomass) exhibited strong diel variations at a depth of one meter, with highest
concentrations during the evening and early morning (18:00-06:00) and much lower values during the day. These findings presumably indicate a strong vertical migratory
behavior of the phytoplankton assemblage.
|
|
|
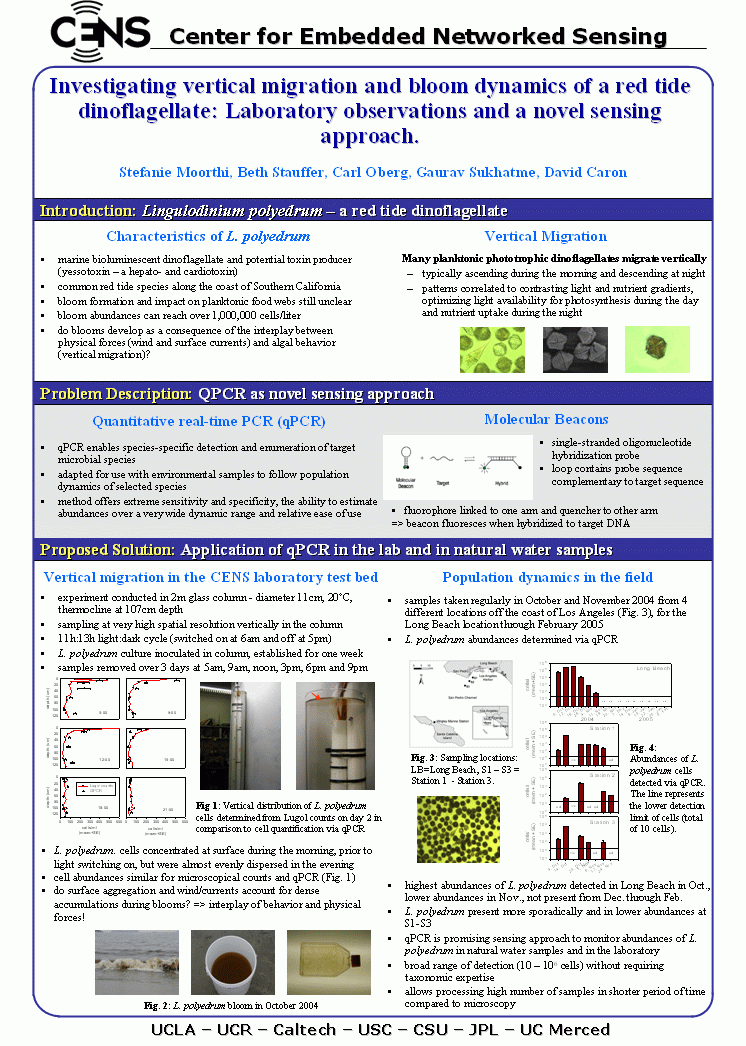
2006 NSF SITE VISIT Poster Session (Jun, 2006)
Investigating vertical migration and bloom dynamics of a red tide dinoflagellate: Laboratory observations and a novel sensing approach ,
Stefanie Moorthi, Beth Stauffer, Carl Oberg, Gaurav Sukhatme, David Caron
Lingulodinium polyedrum is a marine, bioluminescent dinoflagellate that is a common red tide species and potential toxin producer (yessotoxin) along a large expanse
of the coast of Southern California. Little is known about the factors leading to bloom formation, or its impact on planktonic food webs. Bloom abundances can reach
over 1000 cells/ml, events in which the interplay of physical forces (wind and surface currents) and of algal behavior (vertical migration) presumably play an
important role. Using the CENS laboratory test bed, we have examined the pattern and timing of vertical migration by the dinoflagellate in a 2m water column on an
11h:13h light:dark photoperiod. Species-specific detection and enumeration of this organism was accomplished using an 18S rDNA-based quantitative real-time PCR (qPCR)
approach developed in our lab. Positive phototaxis of L. polyedrum resulted in dense aggregations of the dinoflagellate within the top few centimetres of the water
column during the light period, while cells were evenly distributed during the night. The high sensitivity and the broad dynamic range over several orders of
magnitude makes the qPCR sensing method also a very useful new tool for studying the organisms. population dynamics in the field. We subsequently applied the approach
in a study of the seasonal and spatial distribution of L. polyedrum along the coast of Los Angeles. Construction and use of the CENS test bed has allowed a greater
understanding of the vertical migration behavior and its relevance to blooms of this harmful alga.
|
|

|
2006 NSF SITE VISIT Poster Session (Jun, 2006)
NAMOS: Networked Aquatic Microbial Observing System,
Amit Dhariwal, Bin Zhang, Arvind Pereira, Carl Oberg, Beth Stauffer, Stefanie Moorthi, David Caron and Gaurav Sukhatme
As part of our research, we have designed and developed sensor-actuated network for marine monitoring. The network consists of ten static buoy nodes and one mobile
robotic boat for real-time in-situ measurements and analysis of chemical, physical and biologically pertinent phenomena governing the abundance of micro-organisms at
relevant spatio-temporal scales. The goal of the network is to obtain high-resolution information on the spatial and temporal distribution of plankton assemblages in
aquatic environments using the in situ presence afforded by the network, and to make possible network-enabled robotic sampling of hydrographic features of interest.
This work constitutes advances in (1) real-time observing in aquatic ecosystems and (2) sensor-actuated sampling for biological analysis.
Ultimately such systems will be able to establish patterns in the sensed data, and use their mobility to adapt sample collection, a major step forward in the use of
embedded networked sensing in aquatic ecosystems. The field work has demonstrated the basic functionality of the system. The data collected from the deployments
revealed interesting spatio-temporal patterns of chlorophyll, and were useful to validate the design of the buoys and the boat. Static buoys provide low-resolution
spatial sampling with high temporal resolution while a mobile robot boat provides high resolution spatial sampling with low temporal resolution.
|
|

|
2006 International Conference on Robotics and Automation (ICRA 06) (May, 2006)
NAMOS: Networked Aquatic Microbial Observing System,
Amit Dhariwal, Bin Zhang, Beth Stauffer, Carl Oberg, David Caron, Gaurav Sukhatme
|
|
|
|
|
The material presented here is based upon work supported by the
National Science Foundation under Grant No. CCF-0120778. Any
opinions, findings, and conclusions or recommendations expressed
in this material are those of the author(s) and do not necessarily
reflect the views of the National Science Foundation.
|
|







